DNA-encoded chemical libraries are typically selected (“panned”) by affinity capture against virtually any target protein of choice, in analogy with other display technologies (i.e., antibody-phage display).
The DECL library is initially incubated with the target immobilized on a solid support at very low concentration (pM to fM concentrations). Washing cycles separates “bound” compounds from the “unbound”. Subsequently, the DNA-tags of the retained members are PCR-amplified and the amplicon mixture deconvoluted in order to determine the relative concentration of the individual compounds before and after applying the selection pressure. Identified ”hits” are validated in biological/biochemical assays in presence or absence of the DNA-appendage. Eventually, based on collected structural-activity information, a second generation “affinity maturation” library can be generated and used in the next round of selection and decoding (see Video below).
Video: Schematic representation of DECL selection and decoding process. Click on the figure to play the video.
Differently from traditional screening methodologies, in which the biological properties of the individual compound members are discretely determined, selection strategies allows to simultaneously interrogating the entire displayed library imposing the same selection constraints to all library members. In 2010 McGregor et al.reported a novel selection strategy termed interaction-dependent PCR (IDPCR) to perform in solution “one-pot” DECL multiple selections, without the need to immobilize the target protein on a heterogeneous support. In brief, binding between DNA-conjugated targets and DNA-conjugated ligands enables the formation of double stranded DNA-structures otherwise too short to hybridize. Eventually, the hairpin duplex primes the PCR amplification, after the selection process.
In vitro DECL selections can be performed in various conditions. The choice of the most suitable methodology depends on the nature of the target and on the scope of the selection. To date, antigen immobilization onCNBr-activated sepharose resin and His-tag chelation on chromatography resin (IMAC) have been proved to be versatile and efficient method to screen DNA-encoded chemical libraries.
Selection stringency can be strengthen by increasing the number of washing cycles or varying the washing buffer and the coating density of the target antigen. Blocking agents like hearing sperm DNA, bovine serum albumin and detergents (e.g., tween-20) can drastically reduce unspecific binding and enhance the accuracy of the final readout.
As for other display strategies (e.g., phage display), DECL selections can be directed against specific epitope/pocket of the chosen target, either by displacement of a known ligand or by performing multiple panning experiments on a set of structurally related proteins.
qPCR to optimise selection conditions and pre-evaluate selection efficiency
As mentioned in the previous chapter, in a routine DECL selection experiment are typically used 105-108 copies of individual DNA-encoded compounds and the hit-identification is performed comparing the DNA sequencing fingerprints of the library before and after selection. Therefore, quantification of DNA content before and after panning may contribute to provide key information about the stringency and efficiency of the selection conditions, as well as predict the rate of unspecific binding (“stickiness”) of the library on the solid support.
Following qPCR analysis of a DNA-encoded library serial dilution, it is possible to generate PCR-calibration curve, which enable comparative evaluation of DNA recovery in various selection conditions (Figure 18a and b).
Though, theoretically, the detection limit of the qPCR is determined by the amplification of a reaction mixture without template input, which ultimately correspond to the formation of PCR-primer dimers (Figure 18b). In evaluating selection experiments, the dynamic range of qPCR detection is prevalently limited by the selection “noise” due to unspecified binding (“stickiness”) of the library to the support.
Eventually, a preliminary qPCR assessment of the selection output provides a rapid diagnostic test for the quality of the panning experiments and facilitates changes to the experimental conditions, before the DNA sequencing is committed.
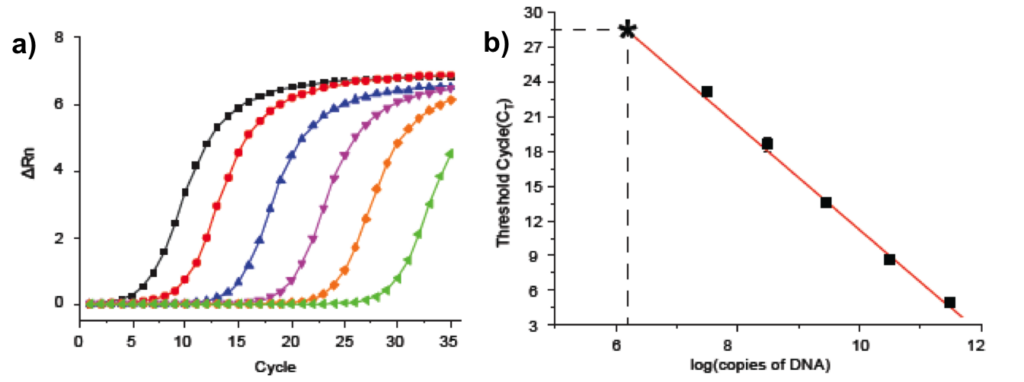
Figure 18: qPCR calibration curve: a) Exemple of qPCR analysis of a DNA-encoded chemical library serial dilution; b) qPCR calibration curve. The asterisk corresponds to amplification of a reaction mixture without template input (i.e., formation of PCR-primer dimers) and ultimately to the theoretical detection limit of the method.

