Cell surface target proteins
In 2015 GSK laboratory described the first cell-based DNA-encoded small-molecule library selection.
Using a protocol very similar to the conventional heterogeneous-phase selection, the DECL library is initially incubated with the cells at 37°C for 30 minutes in 1 mL buffer containing sodium azide to inhibit target internalization.
A DNA-blocking agent, such as sheared salmon sperm DNA, is also included to minimize aspecific binding.
Subsequently, the cells are washed several times (e.g., ten) with ice cold buffer by gentle centrifugation and re-suspension to remove non-binding library members.
Eventually the bound fraction is eluted by heating the cell suspension to 95°C for 10 minutes and collecting the supernatant after high-speed centrifugation of the cell pellet. At this stage, the recovered fraction can be used either in a next round of selection with freshly prepared cells, or for PCR amplification and subsequent deep sequencing decoding.
It was also demonstrated the possibility to use the very same above described protocol in competitive selection, simply adding a fix concentration of a known competitor to the selection buffer at each selection round.
Cell cytosol target proteins
In 2019 the Krusemark research lab at Purdue University, reported the first targeting of DECLs to proteins within the cytosol of living cells.
Using conjugated DNA-linked molecules to a cyclic cell-penetrating peptide (cCPP), featuring high cell entry and endosomal escape efficiency, and applying covalent cross-linking to the library selection, the research group was able to demonstrate the targeting and isolation of nanomolar ligands (i.e., included in the collection as positive controls) from a proof-of-concept DECL-conjugate library.
Cell entry of library members was assessed by fluorescence microscopy and benchmarking of the live cell selection with a cell lysate selection, in absence of cyclic cell-penetrating peptide, revealed a very good correlation of the compound enrichment after sequencing decoding.
In 2021, Vipergen ApS reported the first successful screening of a multimillion membered DECL inside a living cell. Vipergen envisioned the physical injection of a DECL into sufficiently large living cell (i.e., oocytes from Xenopus laevis; >100 000 times bigger than a typical somatic cell) ad-hoc engineered to express in the cytosol the target protein of interest (POI) fused to a “prey” protein (i.e., catalytic domain of human carbonic anhydrase IX) to provide a possible setup for subsequently discriminating DECL members binding to the POI from the ones targeting irrelevant endogenous cell proteins (Figure 1a).
In essence, a mRNA encoding for the POI-“prey” fusion protein is injected into the oocyte, which will then express the protein construct in the cytosol. Subsequently, DECL library together with a “bait”-DNA-conjugate (i.e, a known ligand for catalytic domain of human CAIX reported functional in Xenopus oocytes) is also injected in the oocyte. During incubation, both binding of library members to POI and “bait” to “prey” occur inside the living cell. Eventually, the membrane is broken by centrifugation and the supernatant collected for BTE (binder trap enrichment)mediated DNA ligation and thus submitted to decoding.
Applying such screening architecture (in this regard termed cellular binder trap enrichment, cBTE) to the identification of binders against various pharmaceutical relevant target proteins (i.e., p38α, ACSS2, and DOCK5) it was possible to report the isolation of multiple cluster of ligands with low-micro/nanomolar IC50, in particular towards p38α, including Compound 1 (Figure 1b), which was previously identified by conventional BTE and shown to be functional in a cellular assay with IC50 = 7 nM (Kd = 2.5 nM)
In principle, such conceptual selection architecture (currently demonstrated in combination with BTE) not only enables DEL screening in a living cell under more physiologically relevant conditions, but also eliminate the need for purified active protein, as the selection input is no longer the POI, but the corresponding target gene in the form of mRNA.
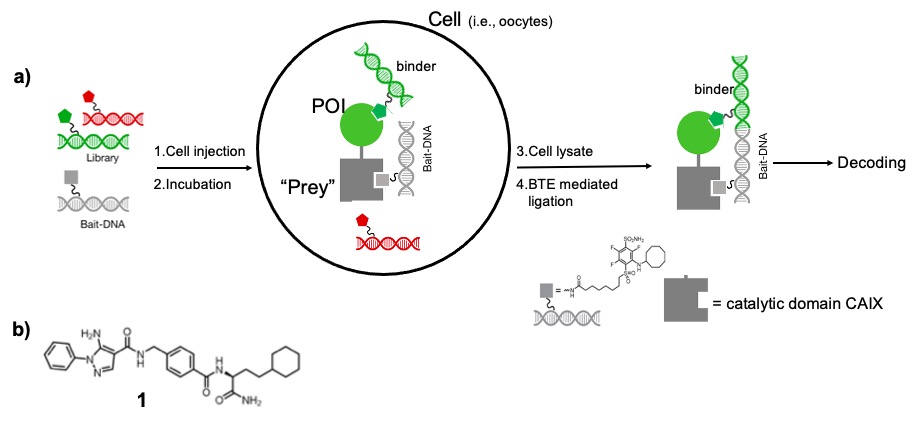
Figure 31: a) cBTE (cellular binder trap enrichment) b) example of p38α ligand: IC50 = 7 nM (cellular assay; Kd = 2.5 nM previously identified as VPC00628)

