In 2004, the group of Prof. Dario Neri at ETH Zurich presented a conceptually different approach for the construction of very large high-quality DNA-encoded chemical libraries.
This strategy, termed Encoded Self-Assembling Chemical (ESAC) library features the non-covalent combinatorial assembly of complementary sub-libraries by means of Watson-Crick DNA-base pairing. Each sub-library consists of oligonucleotides containing both a coding domain, univocally identify the linked chemical moiety (either at the 5’-extremity or at 3’-extremity), and a hybridization domain to a further sub-library pool (Figure 15). In sharp contrast with other DECL technologies, ESAC approach does not involve the formation of stable covalent bonds between different sets of building blocks to generate very complex DNA-encoded libraries.
By means of self-assembly, relatively small ESAC sub-libraries (containing a few thousands of members) can rapidly yield to very large DNA-encoded repertoires (comprising millions of library members), retaining the same initial high quality and purity (Figure 15).
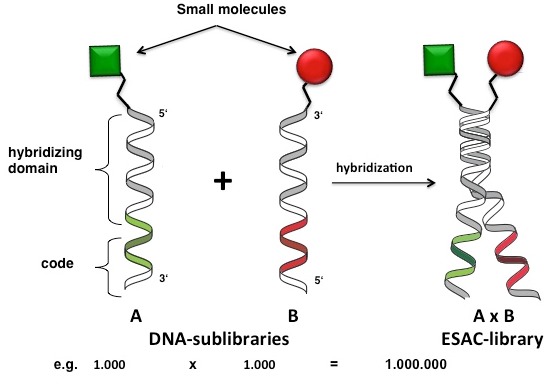
Figure 15: Encoded Self-Assembling Chemical (ESAC) libraries: partially complementary sub-libraries are hybridized by non-covalent Watson-Crick DNA-base pairing. Combinatorial self-assembly of relatively small sub-libraries (i.e., containing 1.000 member) allows the generation of large combinatorial libraries (i.e., comprising 1.000.000 of compound members). Typically, the first sub-library displays chemical entities attached at the 5’-extremity of single-stranded oligonucleotides, whereas the second sub-library are connected through the 3’-extremity to the displayed chemical compound. The display of two neighboring chemical entities may enable the simultaneous engagement of twp adjacent non-overlapping binding sites on the same target protein, in full analogy to fragment-based drug discovery approaches. Reproduced by permission of The Royal Society of Chemistry (http://pubs.rsc.org/en/content/articlelanding/2011/cc/c1cc15634a).
In sharp contrast with other DECL technologies, ESAC approach does not involve the formation of stable covalent bonds between different sets of building blocks to generate very complex DNA-encoded libraries.Indeed,library members are individually synthesized and HPLC purified, thus ensuring elevated purity and about 100% display for all library conjugates. As shown in Figure 16, ESAC technology can be implemented in at least three different embodiments. In a first setup each sub-library is independently used for affinity capture and selection experiments (Figure 16a). Differently, in lead optimization mode, a ESAC sub-library is annealed with a single oligonucleotide displaying a known binder to a target protein, in order to perform “affinity maturation” of the specific compound (Figure 16b). Eventually, if two (or three) independent ESAC sub-sets are hybridized, the resulting combinatorial hetero-douplex (or hetero-triplex) library can be employed in the selection for the de novo discovery of bi/tridentate binding compounds against the target of choice (Figure 16c).
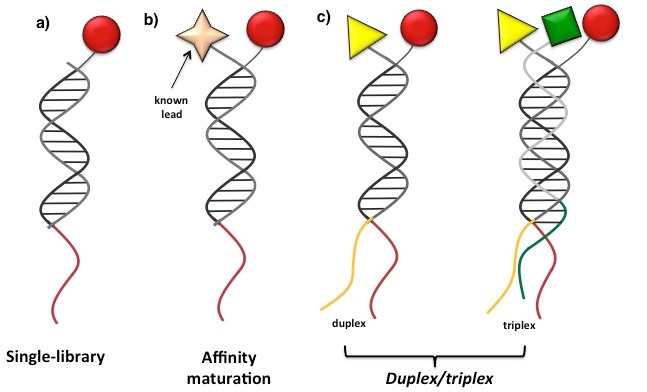
Figure 16: ESAC t echnology embodiments: a) “single-library” format; b) “affinity maturation” setting; c) hetero-douplex (or hetero-triplex) library for the de novo discovery of binding molecules. Reproduced by permission of The Royal Society of Chemistry (http://pubs.rsc.org/enarticlelanding/2011/cc/ /c1cc15634a).
In analogy with traditional fragment-based approach, at the end of the selection process, the newly identified binding pairs must be connected into a single drug-like high affinity compound. Although the determination of the optimal linkage (in terms oflength, flexibility and position) by combinatorial medicinal chemistry can be relatively cumbersome, Neri and co-workers successfully used ESAC technology in various discovery campaigns, including the de novoidentification of binders as well as the improvement of previously discovered lead structures.
In 2011, Winssinger and co-workers have developed a conceptually similar strategy to ESAC technology,based on the combinatorial self-assembly of two PNA-libraries on single-stranded DNA-templates (Figure 17). In principle, all the chemistry can be performed in automated fashion (using a peptide synthesizer for the synthesis of the PNA conjuates and an oligonucleotide synthesizer for the DNA-templates). However, for the theoretical construction of a one million member library (1.000 x 1.000 library compounds), it would require the bulky and costly synthesis of 2.000 PNA-conjugates as well as a tedious enzymatic assembling of 1 million different DNA-templates. Nonetheless, the strategy as presented lends itself to the “molecular evolution” of its library members, a feature not yetanticipated by ESAC technology(Figure 17).
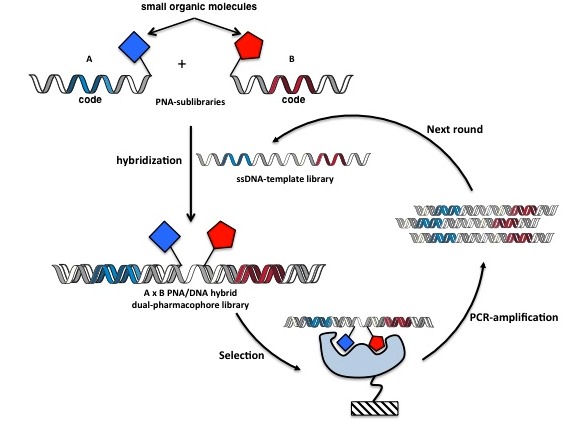
Figure 17: PNA/DNA self-assembly encoded library. Two independent PNA-sublibraries each displaying a chemical moiety and a compound specific coding domain hybridize on a complementary DNA-template. Following selection of the resulting library and PCR amplification of the enriched DNA-tags, dsDNA is converted into ssDNA. Next-generation library is reassembled re-exposing the templates to PNA-sublibrary fragments.

