In 2004 Pehr Harbury and David Halpin developed a variation of the“pool-split-pool” strategy applied to the construction of DNA-conjugate libraries.
In this alternative approach, the DNA-appendage serves both as a PCR-amplifiable identification tag as well as code for programming the stepwise library assembly.
As illustrated in Figure 9, the individual oligonucleotide templates are “routed” (split) into sequential cartridges containing specific anti-codon DNA sequences. Each template is processed in a discreet chemical reaction (coupling), eluted (pool) and subsequently flown through a further set of anti-codon columns until all the desired building blocks are incorporated.
In a first practical implementation Halpin and coworkers chemically translated 106 different 340-mer-oligonucleotide templates, each containing six coding regions [(a-j)1-6] and seven constant domains (z1-7), into corresponding 106 N-acylated pentapeptides (Figure 9). Sequencing of the resulting library before and after two rounds of selection and chemical translation against a anti-luecine-enkephalin antibody revealed a remarkable enrichment of the expected consensus pentapetidic sequences, intentionally included during the synthesis as positive controls.
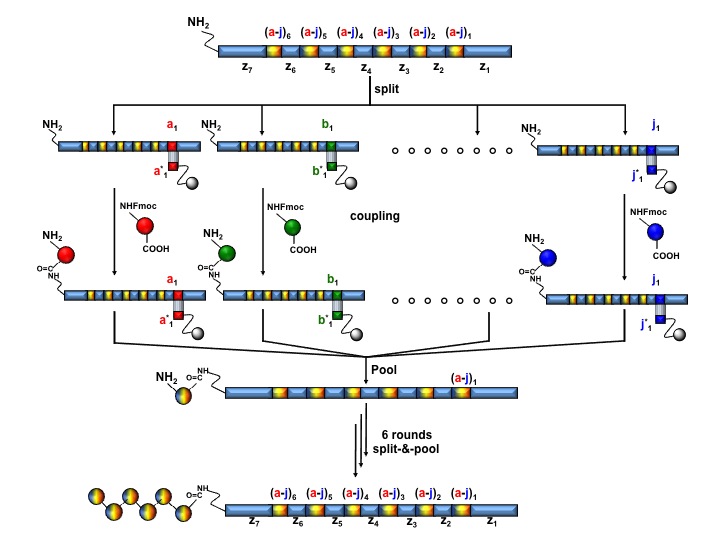
Figure 9: Schematic representation of a DNA-routed library assemply comprising 106 N-acylated pentapeptides.
It worth noting that as the DNA-appendage serves both for encoding as well as for programming the library assembly, the templates surviving the selection process could be used as “evolutionary” input in further rounds of chemical translation and selection. In this regards, this work represents the first demonstration of “molecular evolution” by means of a DNA-encoded chemical library.
Although the chemical translation of DNA-templates by “DNA-routing” holds promises for the construction of large libraries whose sizes increase exponentially after each round of synthesis, from a drug-discovery point of view, the simple polypeptidic structures which have been so far assembled using such methodology may not represent the drug-like structures that pharmaceutical industry is traditionally interested in.

